 |
Xiao WangMember of Technical Staff@xAI |
About Me
|
Xiao Wang is currently a Member of Technical Staff of xAI, focusing on pre-training of vision and multimodal. Prior to that, he obtained a CS Ph.D. degree from Purdue University, focusing on representation learning and AI for biology. He graduated with a bachelor's degree in CS from Xi'an Jiaotong University, Xi'an, China. He used to work at TikTok AI, the University of Washington, Futurewei AI Lab, JD AI Research, and Meta AI (FAIR Labs). His research interests include pre-training of multimodal, representation learning, generative modeling and AI for biology. |
Fellowship&Awards
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Selected publications
* denotes equal contribution2025
|
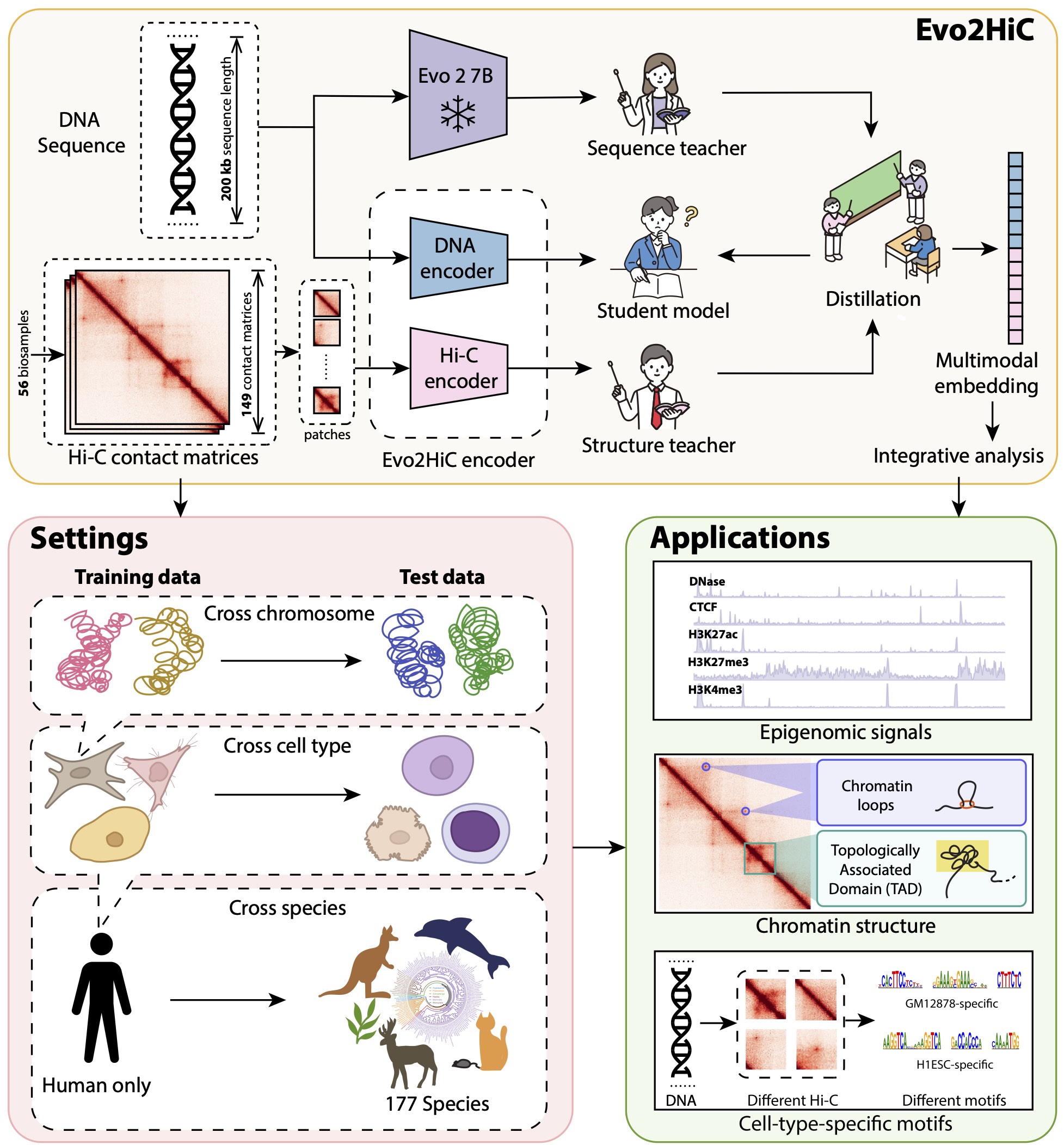
Evo2HiC: a multimodal foundation model for integrative analysis of genome sequence and architecture Tangqi Fang* Xiao Wang*, Zhiping Xiao, Shengqi Hang, Ghulam Murtaza, Junwei Yang, Hanwen Xu, Anupama Jha, William Noble, Sheng Wang Nature Genetics (under review), 2025 Paper Code [AI for biology], [Representation learning] |
|
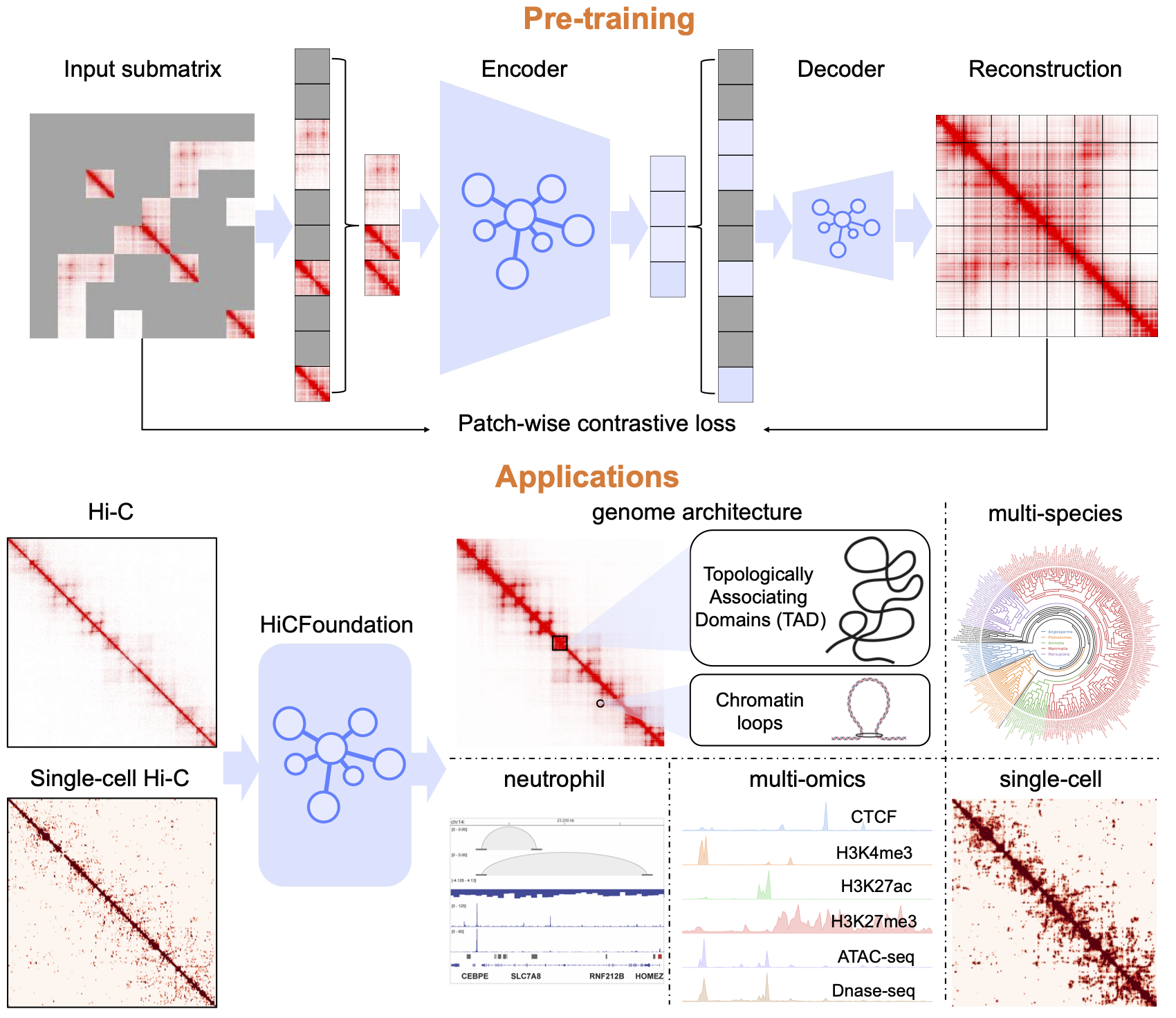
A generalizable foundation model for chromatin architecture, single-cell and multi-omics analysis across species Xiao Wang*, Yuanyuan Zhang*, Suhita Ray, Anupama Jha, Tangqi Fang, Shengqi Hang, Sergei Doulatov, William Stafford Noble, Sheng Wang Nature Methods (accepted), 2025 Paper Code Colab [AI for biology], [Generative modeling], [Representation learning] |
|
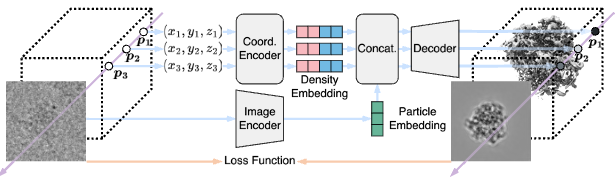
CryoNeRF: generalizable automated cryo-EM reconstruction using neural radiance field Huaizhi Qu*, Xiao Wang*, Yuanyuan Zhang, Sheng Wang, William Stafford Noble, Tianlong Chen BioRxiv, 2025 Paper Code [AI for biology], [Representation learning] |
|
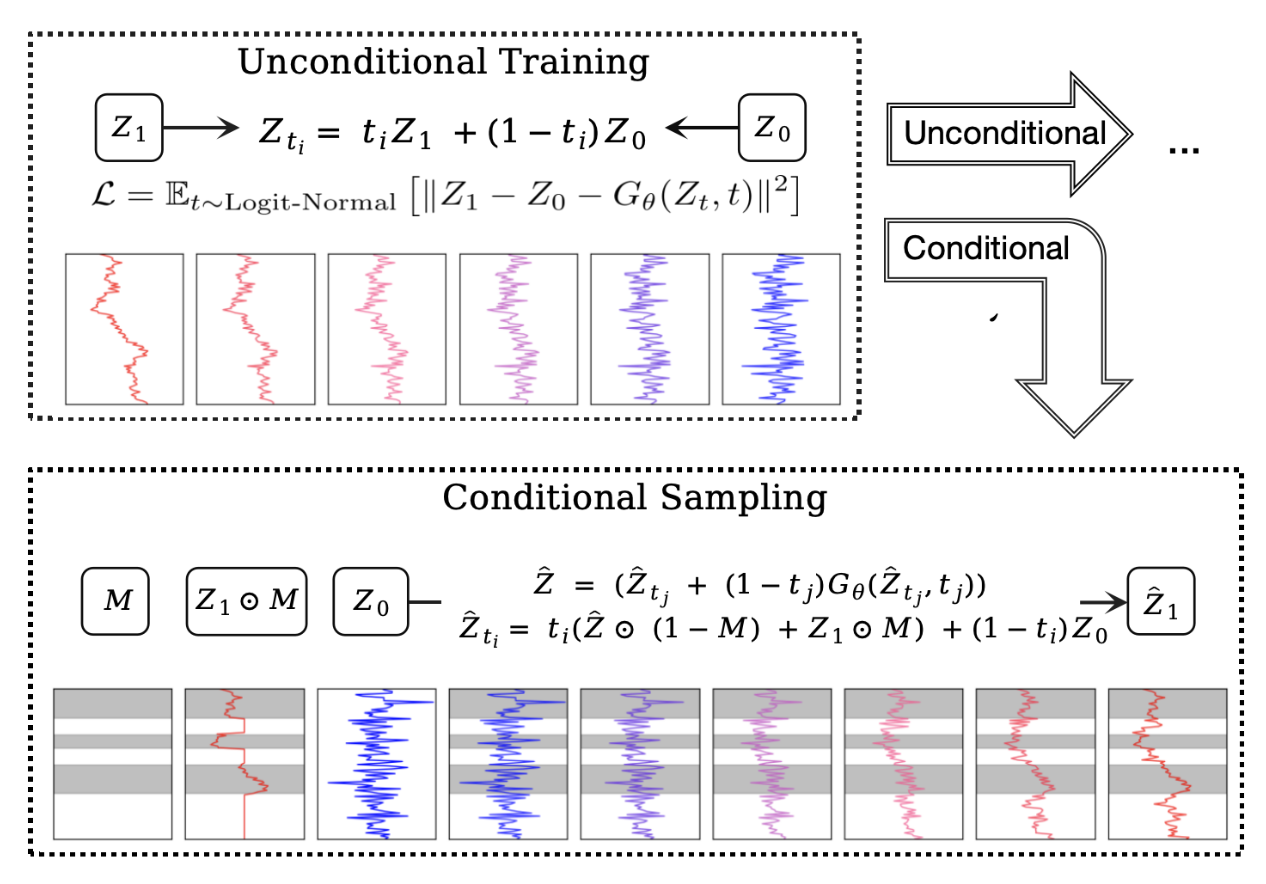
FlowTS: Time Series Generation via Rectified Flow Yang Hu*, Xiao Wang*, Zezhen Ding, Lirong Wu, Huatian Zhang, Stan Z. Li, Sheng Wang, Jiheng Zhang, Ziyun Li, Tianlong Chen Arxiv, 2025 Paper Code [Generative modeling] |
2024

|
DiffModeler: Large Macromolecular Structure Modeling for Cryo-EM Maps Using Diffusion Model Xiao Wang, Han Zhu, Genki Terash, Manav Taluja, Daisuke Kihara Nature Methods, 2024 Paper Code Server [AI for biology], [Generative modeling] |
|
DeepMainmast: integrated protocol of protein structure modeling for cryo-EM with deep learning and structure prediction Genki Terash, Xiao Wang, Devashish Prasad, Tsukasa Nakamura, Daisuke Kihara Nature Methods, 2024 Paper Code Colab Server Selected as the cover of Volume 21 issue 1 of Nature Methods [AI for biology] |
2023
|
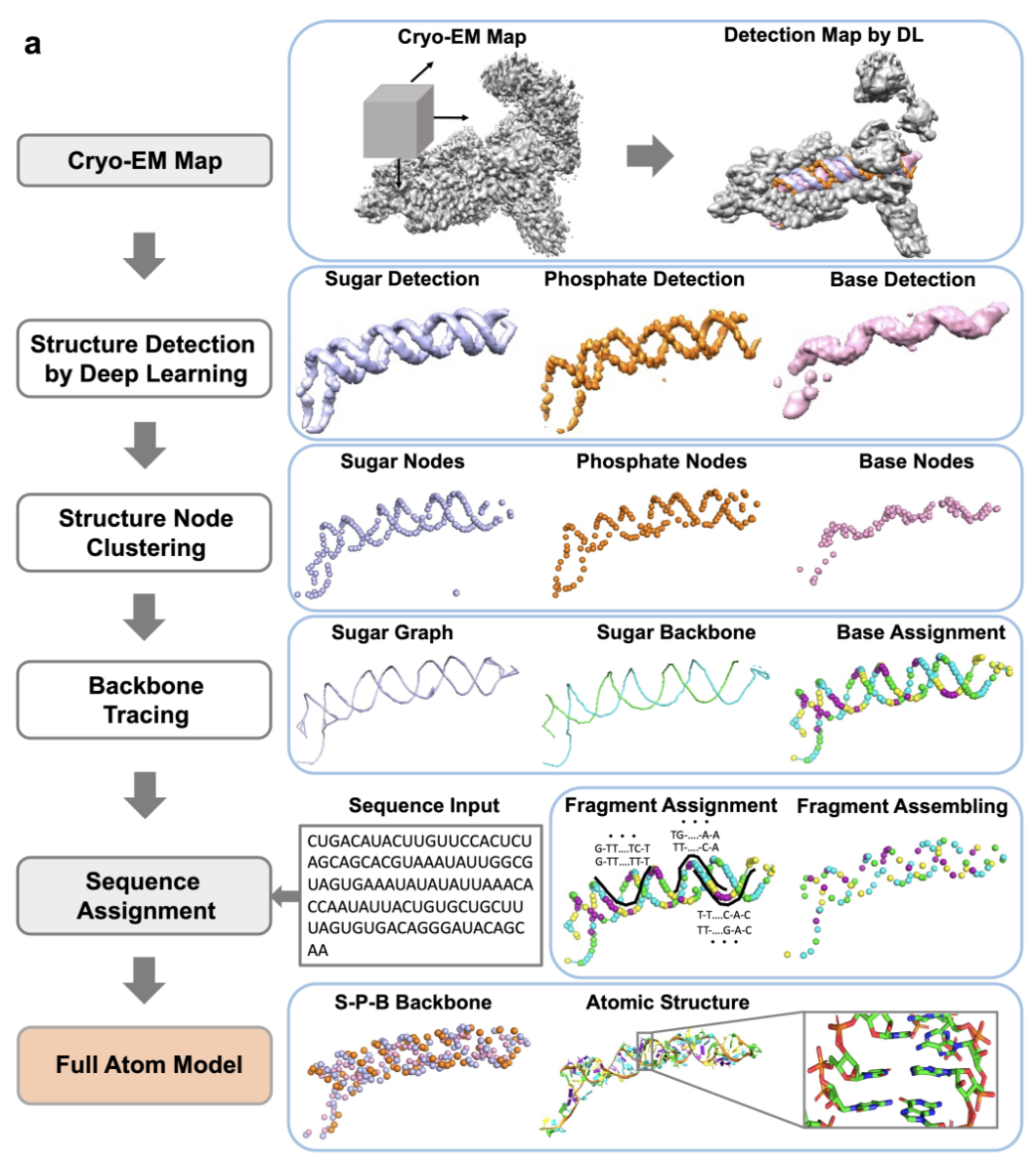
De Novo Structure Modeling for Nucleic Acids in cryo-EM Maps Using Deep Learning Xiao Wang, Genki Terash, Daisuke Kihara Nature Methods, 2023 Paper Code Colab Server Selected for research briefing CryoREAD provides fully automated DNA–RNA structure modeling for cryo-EM maps in Nature Methods. [AI for biology] |
|
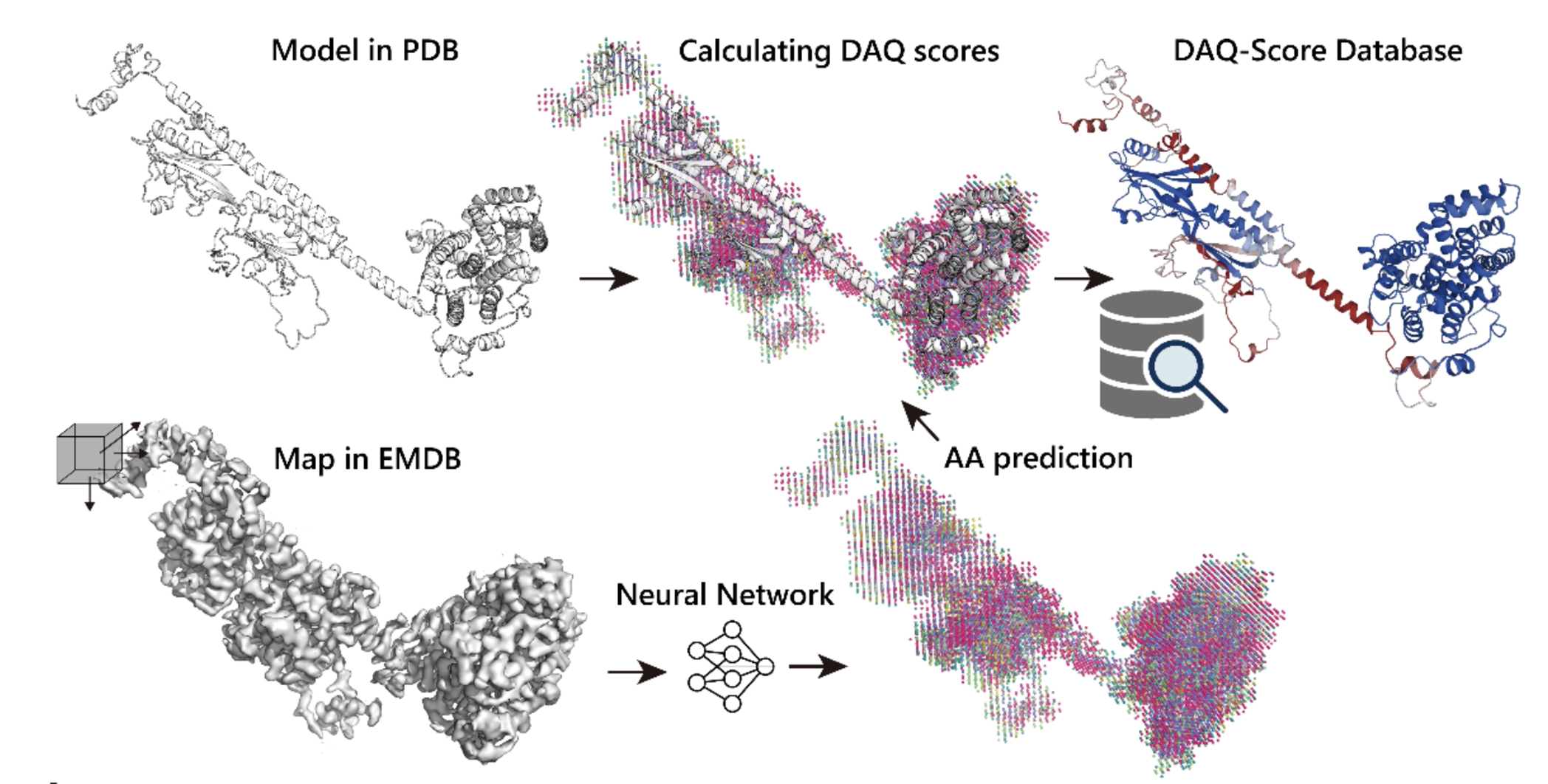
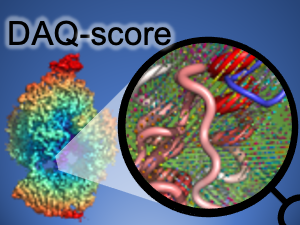
DAQ-Score Database: Assessment of Map-Model Compatibility for Protein Structure Models from Cryo-EM Maps Tsukasa Nakamura, Xiao Wang, Genki Terash, Daisuke Kihara Nature Methods, 2023 Paper Database Server [AI for biology] |
|
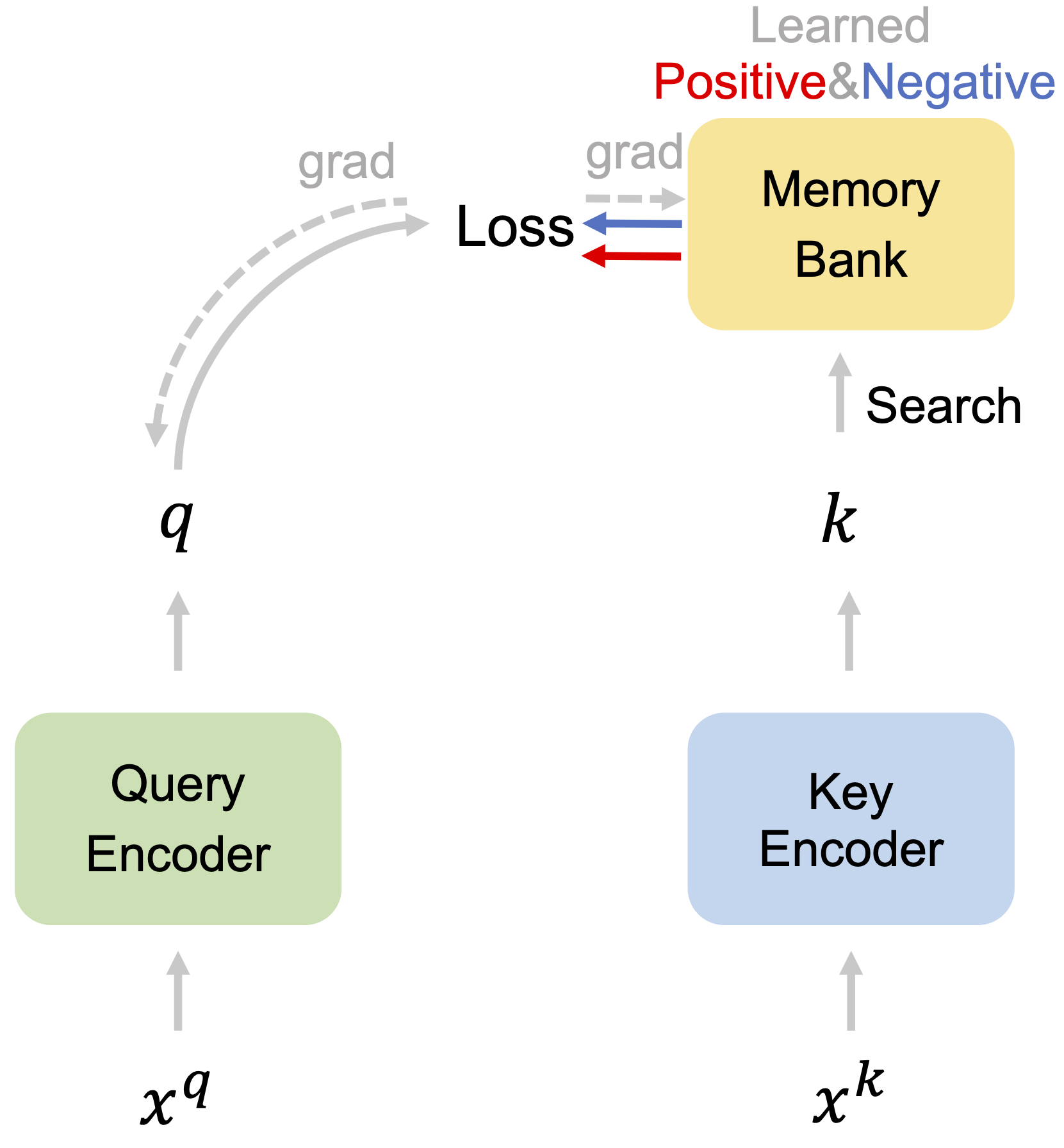
CaCo: Both Positive and Negative Samples are Directly Learnable via Cooperative-adversarial Contrastive Learning Xiao Wang, Yuhang Huang, Dan Zeng, Guo-Jun Qi IEEE Transactions on Pattern Analysis and Machine Intelligence (IEEE T-PAMI), 2023 Paper Code [Representation learning] |
2022
|
Residue-Wise Local Quality Estimation for Protein Models from Cryo-EM Maps Genki Terash*, Xiao Wang*, Sai Raghavendra Maddhuri Venkata Subramaniya, John J. G. Tesmer, Daisuke Kihara Nature Methods, 2022 Paper Code Colab Server [AI for biology] |
|
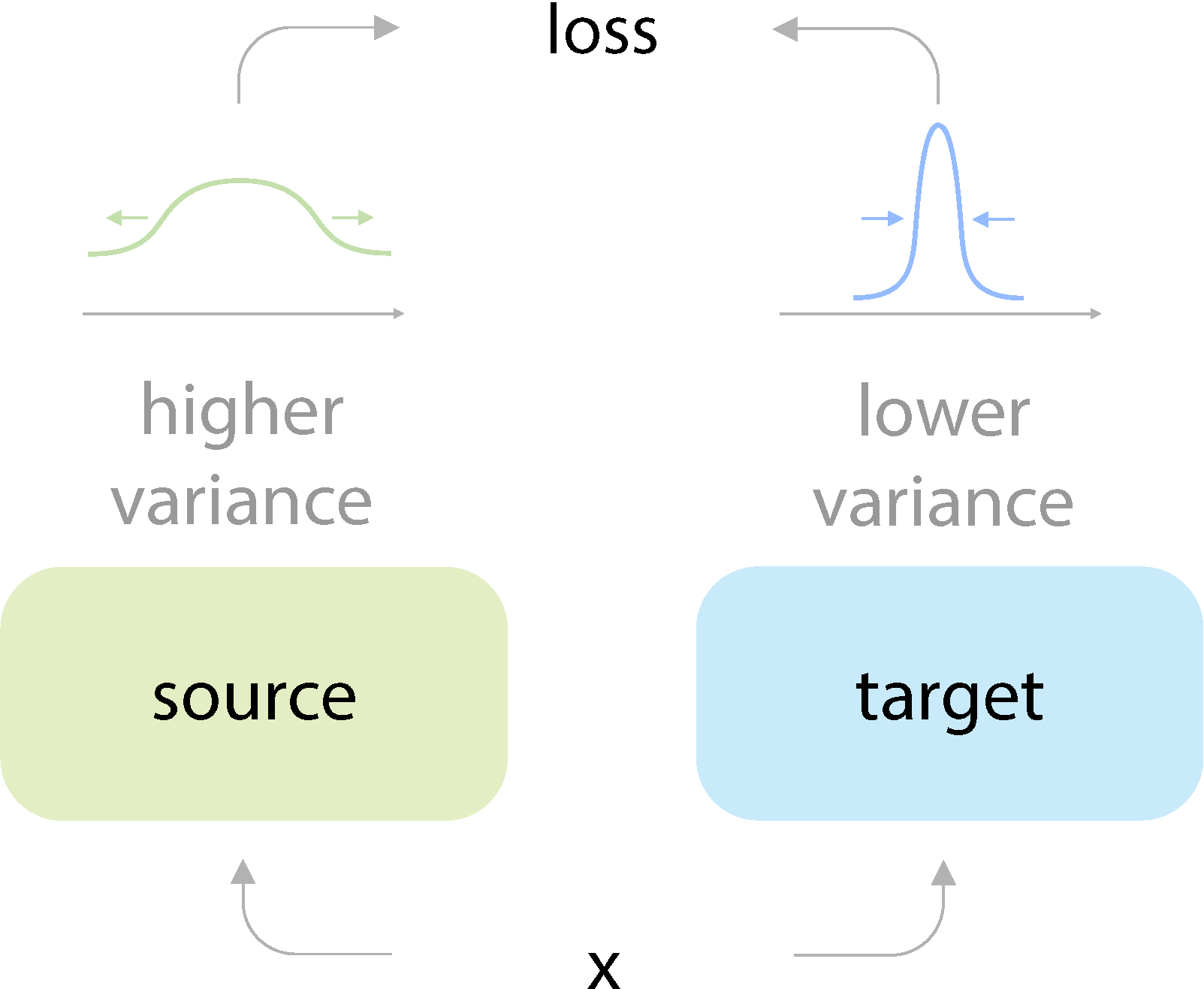
On the Importance of Asymmetry for Siamese Representation Learning Xiao Wang*, Haoqi Fan*, Yuandong Tian, Daisuke Kihara, Xinlei Chen Conference on Computer Vision and Pattern Recognition (CVPR), 2022 Paper Code [Representation learning] |
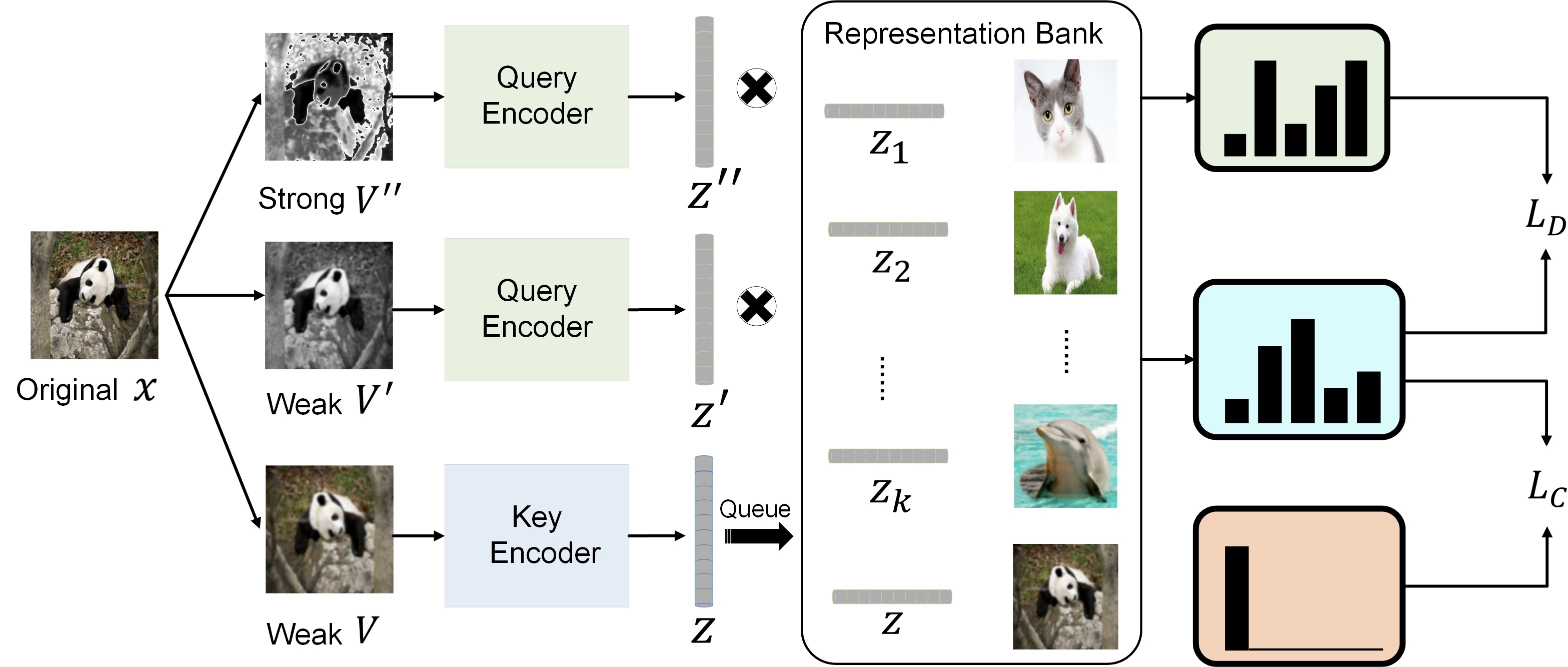 |
Contrastive Learning with Stronger Augmentation Xiao Wang, Guo-Jun Qi IEEE Transactions on Pattern Analysis and Machine Intelligence (IEEE T-PAMI), 2022 Paper Code [Representation learning] |
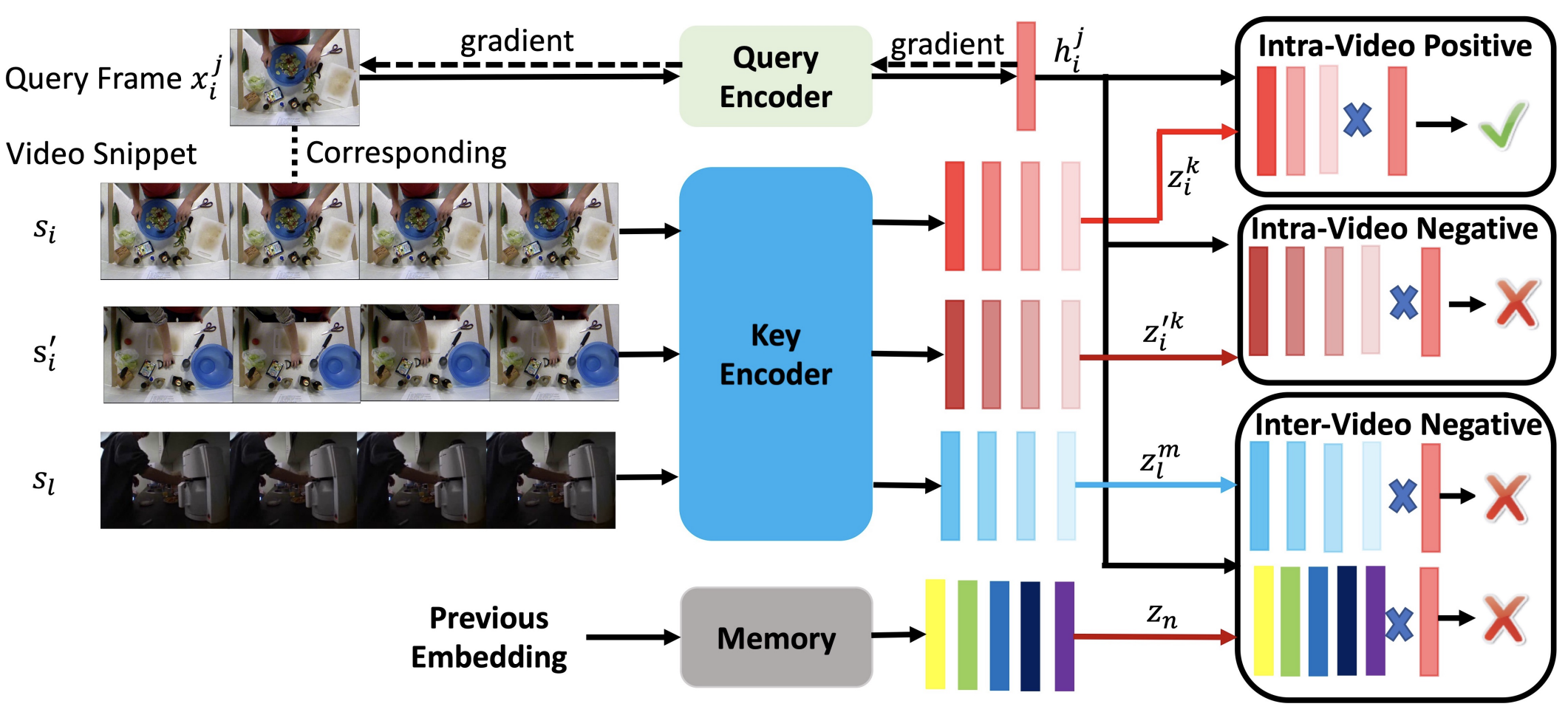 |
CoSeg: Cognitively Inspired Unsupervised Generic Event Segmentation Xiao Wang, Jingen Liu, Tao Mei, Jiebo Luo IEEE Transactions on Neural Networks AND Learning Systems (IEEE TNNLS), 2022 Paper code [Representation learning] |
2021
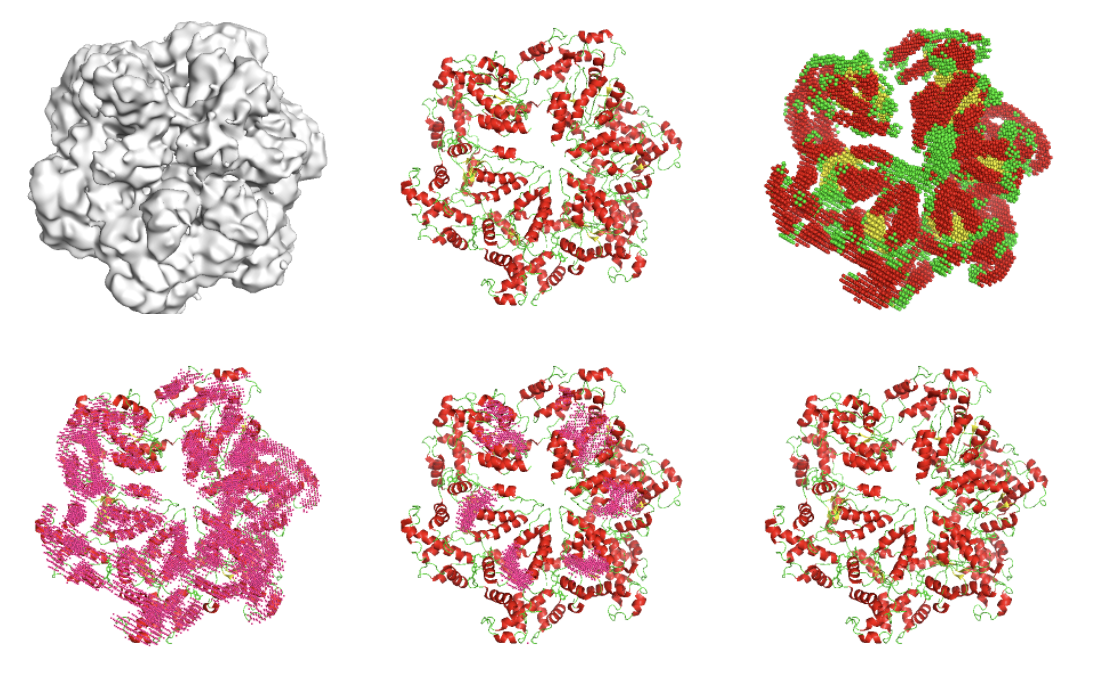 |
Detecting Protein and DNA/RNA Structures in Cryo-EM Maps of Intermediate Resolution Using Deep Learning Xiao Wang, Eman Alnabati, Tunde W Aderinwale, Sai Raghavendra Maddhuri, Genki Terashi, Daisuke Kihara Nature Communications, 2021 Paper Code Code_Ocean Colab Server [AI for biology] |
|
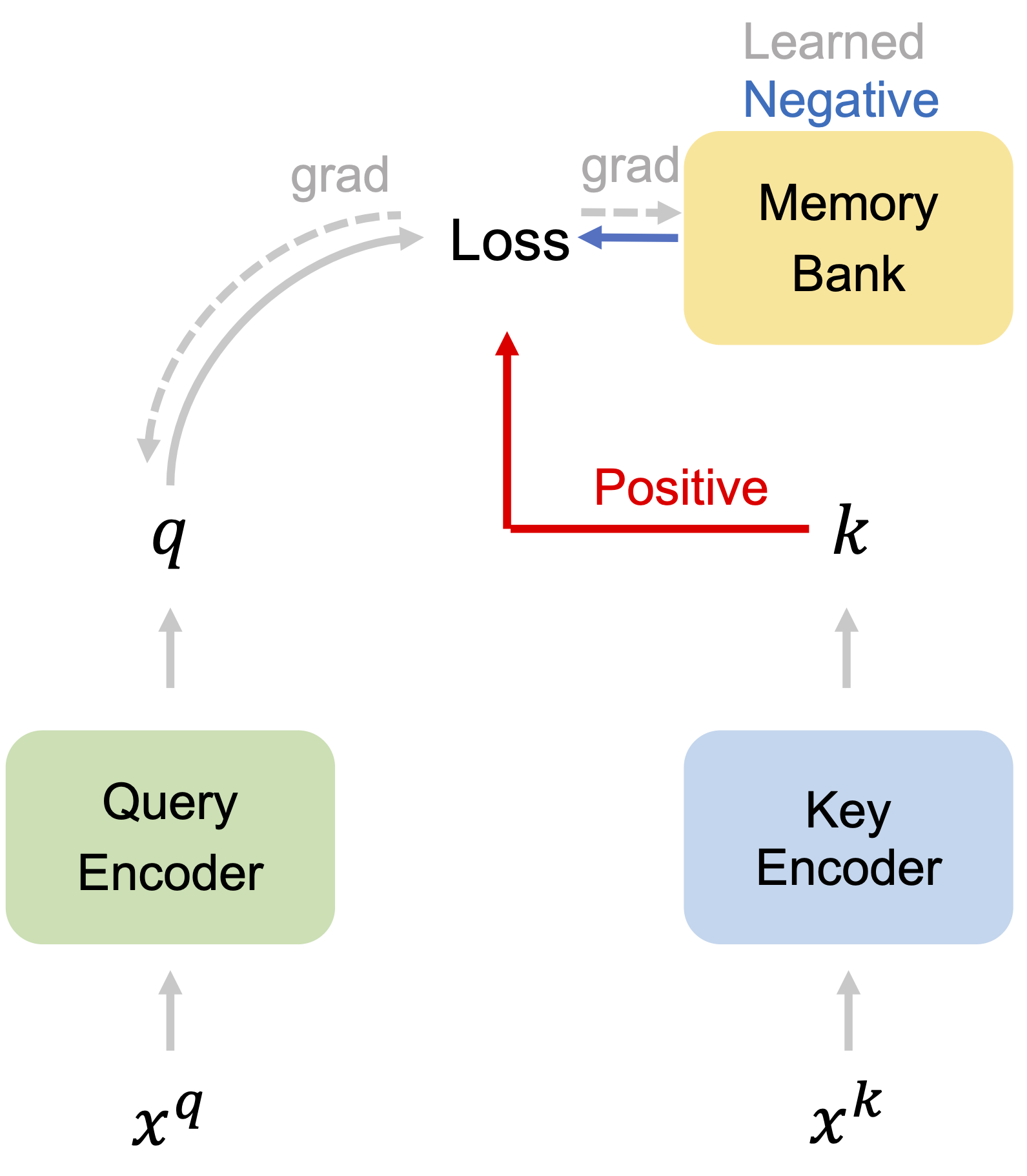
Adco: Adversarial contrast for efficient learning of unsupervised representations from self-trained negative adversaries Xiao Wang*, Qianjiang Hu*, Wei Hu, Guo-Jun Qi Conference on Computer Vision and Pattern Recognition (CVPR), 2021 Paper Code [Representation learning] |
2020
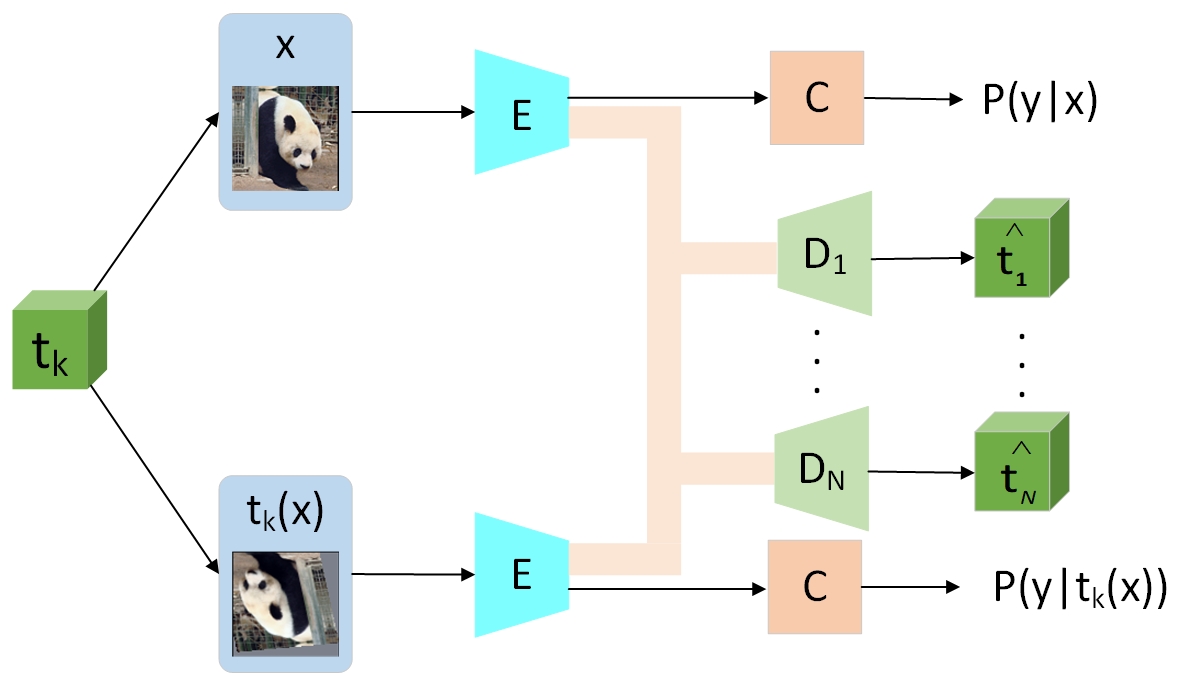 |
EnAET: Self-Trained Ensemble AutoEncoding Transformations for Semi-Supervised Learning Xiao Wang, Daisuke Kihara, Jiebo Luo, Guo-Jun Qi IEEE Transactions on Image Processing (IEEE TIP) , 2020 Paper code Learning generalized transformation equivariant representations via autoencoding transformations Guo-Jun Qi, Liheng Zhang, Feng Lin, Xiao Wang IEEE Transactions on Pattern Analysis and Machine Intelligence (IEEE T-PAMI), 2020 Paper code [Representation learning] |





















